Microvolution® software delivers nearly instantaneous deconvolution by combining intelligent software programming with the power of a GPU
Developed by Stanford scientists, Microvolution software will improve your research:
- Work more effectively in dim light and realize greater success rates with live-cell and time-lapse experiments
- Create cleaner measurements after deconvolution (e.g., colocalization, FRET data, neurite lengths, fluorescence intensities)
- Make adjustments to your microscopy experiments on the fly and collect more data from the same sample
The software deconvolves images from widefield, confocal, two photon, light sheet, and HCA microscopes. Blind deconvolution option improves noisy data, such as deep tissue imaging. Multi-GPU options enable even giant images to be processed in seconds.
*Bruce MA, Butte MJ. Real-time GPU-based 3D Deconvolution, Optics Express, 2013; 21(4): 4766.
Speed
Deconvolve your fluorescence images, from small to very large, in a fraction of the time
Accuracy
Thin Filaments Are Preserved with Microvolution Software
Microvolution’s method starts with the proven Richardson-Lucy algorithm that is used by most software programs. Other vendors take mathematical shortcuts to speed up iterations, resulting in imprecise images after deconvolution. Microvolution takes no shortcuts. Our software delivers accurate images, up to 200 times faster.
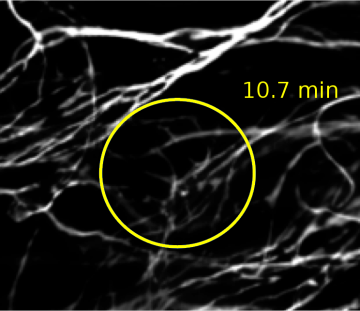

Image courtesy of Molecular Devices.
Clarity
Increased resolution
When collected under the right conditions, deconvolution can help break the diffraction barrier. Pictured below are 180 nm separated lines on an Argo-SIM slide, imaged with widefield microscopy. Deconvolution brings a √2 improvement in visual resolution.


Image courtesy of Tong Zhang and Puifai Santisakultarm, Salk Institute.
Citations
Microvolution has been cited in the following publications
| Gudmundsson, S. R. et al. Morphology of Phagophore Precursors by Correlative Light-Electron Microscopy. Cells 11, 3080 (2022) doi:10.3390/cells11193080 |
| Puckett, E. E. et al. Genetic architecture and evolution of color variation in American black bears. (2022) doi:10.1101/2022.08.12.503788 |
| Chen, B. et al. Resolution doubling in light-sheet microscopy via oblique plane structured illumination. (2022) doi:10.1101/2022.05.19.492671 |
| Niide, T., Asari, S., Kawabata, K. & Hara, Y. Specificity of Nuclear Size Scaling in Frog Erythrocytes. Front. Cell Dev. Biol. 10, (2022) doi:10.3389/fcell.2022.857862 |
| Birnbaum, F., Eguchi, A., Pardon, G., Chang, A. C. Y. & Blau, H. M. Tamoxifen treatment ameliorates contractile dysfunction of Duchenne muscular dystrophy stem cell-derived cardiomyocytes on bioengineered substrates. npj Regen Med 7, (2022) doi:10.1038/s41536-022-00214-x |
| Zhang, W. et al. Identification of cell types in multiplexed in situ images by combining protein expression and spatial information using CELESTA reveals novel spatial biology. (2022) doi:10.1101/2022.02.02.478888 |
| Zimmer, S. E. et al. Differential Pathomechanisms of Desmoglein 1 Transmembrane Domain Mutations in Skin Disease. Journal of Investigative Dermatology 142, 323-332.e8 (2022) doi:10.1016/j.jid.2021.07.154 |
| Zhovmer, A. S. et al. Mechanical Counterbalance of Kinesin and Dynein Motors in a Microtubular Network Regulates Cell Mechanics, 3D Architecture, and Mechanosensing. ACS Nano (2021) doi:10.1021/acsnano.1c04435 |
| Ma, Z. et al. Single-cell transcriptomics reveals a conserved metaplasia program in pancreatic injury. Gastroenterology (2021) doi:10.1053/j.gastro.2021.10.027 |
| Jones, N. G. et al. Bromodomain factor 5 is an essential transcriptional regulator of the Leishmania genome. (2021) doi:10.1101/2021.09.29.462384 |
| Hülsemann, M. et al. TC10 regulates breast cancer invasion and metastasis by controlling membrane type-1 matrix metalloproteinase at invadopodia. Commun Biol 4, (2021) doi:10.1038/s42003-021-02583-3 |
| Lucas, R. M. et al. SCIMP is a spatiotemporal transmembrane scaffold for Erk1/2 to direct pro-inflammatory signaling in TLR-activated macrophages. Cell Reports 36, 109662 (2021) doi:10.1016/j.celrep.2021.109662 |
| Lefevre, J. G. et al. LLAMA: a robust and scalable machine learning pipeline for analysis of large scale 4D microscopy data: analysis of cell ruffles and filopodia. BMC Bioinformatics 22, (2021) doi:10.1186/s12859-021-04324-z |
| Thillaiappan, N. B., Smith, H. A., Atakpa-Adaji, P. & Taylor, C. W. KRAP tethers IP3 receptors to actin and licenses them to evoke cytosolic Ca2+ signals. Nat Commun 12, (2021) doi:10.1038/s41467-021-24739-9 |
| Geoghegan, V., Jones, N. G., Dowle, A. & Mottram, J. C. Protein kinase signalling at the Leishmania kinetochore captured by XL-BioID. (2021) doi:10.1101/2021.07.08.451598 |
| Fu, G. et al. Metabolic control of TFH cells and humoral immunity by phosphatidylethanolamine. Nature 595, 724–729 (2021) doi:10.1038/s41586-021-03692-z |
| Silva, L. M., Moutsopoulos, N., Bugge, T. H. & Doyle, A. Live Imaging and Quantification of Neutrophil Extracellular Trap Formation. Current Protocols 1, (2021) doi:10.1002/cpz1.157 |
| Tabdanov, E. D. et al. Engineering T cells to enhance 3D migration through structurally and mechanically complex tumor microenvironments. Nat Commun 12, (2021) doi:10.1038/s41467-021-22985-5 |
| Bowman, S. L. et al. A BLOC-1–AP-3 super-complex sorts a cis-SNARE complex into endosome-derived tubular transport carriers. Journal of Cell Biology 220, (2021) doi:10.1083/jcb.202005173 |
| Shin, G. J. et al. Integrins protect sensory neurons in models of paclitaxel-induced peripheral sensory neuropathy. Proc Natl Acad Sci USA 118, e2006050118 (2021) doi:10.1073/pnas.2006050118 |
| Baxter, B. D. et al. Transcriptional profiling reveals potential involvement of microvillous TRPM5-expressing cells in viral infection of the olfactory epithelium. BMC Genomics 22, (2021) doi:10.1186/s12864-021-07528-y |
| Shimogama, S., Iwao, Y. & Hara, Y. Yolk platelets impede nuclear expansion in Xenopus embryos. (2021) doi:10.1101/2021.01.13.426473 |
| Zhovmer, A. & Combs, C. A. A Step-by-Step Guide to Instant Structured Illumination Microscopy (iSIM). in Confocal Microscopy 347–359 (Springer US, 2021). doi:10.1007/978-1-0716-1402-0_19 |
| Heijo, H. et al. DNA content contributes to nuclear size control in Xenopus laevis. Molecular Biology of the Cell 31, 2703–2717 (2020) doi:10.1091/mbc.e20-02-0113 |
| Le, L. et al. SLC45A2 protein stability and regulation of melanosome pH determine melanocyte pigmentation. Molecular Biology of the Cell 31, 2687–2702 (2020) doi:10.1091/mbc.e20-03-0200 |
| FitzGerald, L. I. et al. A molecular sensor to quantify the localization of proteins, DNA and nanoparticles in cells. Nat Commun 11, (2020) doi:10.1038/s41467-020-18082-8 |
| Schürch, C. M. et al. Coordinated Cellular Neighborhoods Orchestrate Antitumoral Immunity at the Colorectal Cancer Invasive Front. Cell 182, 1341-1359.e19 (2020) doi:10.1016/j.cell.2020.07.005 |
| da Silva, J. L., Gupta, S., Olivier, K. N. & Zelazny, A. M. Antimicrobial peptides against drug resistant Mycobacterium abscessus. Research in Microbiology 171, 211–214 (2020) doi:10.1016/j.resmic.2020.03.001 |
| Pennamen, P. et al. BLOC1S5 pathogenic variants cause a new type of Hermansky–Pudlak syndrome. Genetics in Medicine 22, 1613–1622 (2020) doi:10.1038/s41436-020-0867-5 |
| Meng, K. P., Majedi, F. S., Thauland, T. J. & Butte, M. J. Mechanosensing through YAP controls T cell activation and metabolism. Journal of Experimental Medicine 217, (2020) doi:10.1084/jem.20200053 |
| Chen, Z. et al. Mitochondrial DNA segregation and replication restrict the transmission of detrimental mutation. Journal of Cell Biology 219, (2020) doi:10.1083/jcb.201905160 |
| Chen, X., Wang, K., Svitkina, T. & Bi, E. Critical Roles of a RhoGEF-Anillin Module in Septin Architectural Remodeling during Cytokinesis. Current Biology 30, 1477-1490.e3 (2020) doi:10.1016/j.cub.2020.02.023 |
| Rajakylä, E. K. et al. Assembly of Peripheral Actomyosin Bundles in Epithelial Cells Is Dependent on the CaMKK2/AMPK Pathway. Cell Reports 30, 4266-4280.e4 (2020) doi:10.1016/j.celrep.2020.02.096 |
| Hoffman, D. P. et al. Correlative three-dimensional super-resolution and block-face electron microscopy of whole vitreously frozen cells. Science 367, eaaz5357 (2020) doi:10.1126/science.aaz5357 |
| Jang, S.-M. et al. The RepID–CRL4 ubiquitin ligase complex regulates metaphase to anaphase transition via BUB3 degradation. Nature Communications 11, (2020) doi:10.1038/s41467-019-13808-9 |
| Jacobo, A., Dasgupta, A., Erzberger, A., Siletti, K. & Hudspeth, A. J. Notch-Mediated Determination of Hair-Bundle Polarity in Mechanosensory Hair Cells of the Zebrafish Lateral Line. Current Biology 29, 3579-3587.e7 (2019) doi:10.1016/j.cub.2019.08.060 |
| Chakraborty, T. et al. Light-sheet microscopy of cleared tissues with isotropic, subcellular resolution. Nature Methods 16, 1109–1113 (2019) doi:10.1038/s41592-019-0615-4 |
| Kim, C., Seedorf, G. J., Abman, S. H. & Shepherd, D. P. Heterogeneous response of endothelial cells to insulin-like growth factor 1 treatment is explained by spatially clustered sub-populations. Biology Open 8, bio045906 (2019) doi:10.1242/bio.045906 |
| Sancer, G. et al. Modality-Specific Circuits for Skylight Orientation in the Fly Visual System. Current Biology 29, 2812-2825.e4 (2019) doi:10.1016/j.cub.2019.07.020 |
| Özel, M. N. et al. Serial Synapse Formation through Filopodial Competition for Synaptic Seeding Factors. Developmental Cell 50, 447-461.e8 (2019) doi:10.1016/j.devcel.2019.06.014 |
| Vogt, E.-J. et al. Anchoring cortical granules in the cortex ensures trafficking to the plasma membrane for post-fertilization exocytosis. Nature Communications 10, (2019) doi:10.1038/s41467-019-10171-7 |
| Mohan, A. S. et al. Enhanced Dendritic Actin Network Formation in Extended Lamellipodia Drives Proliferation in Growth-Challenged Rac1P29S Melanoma Cells. Developmental Cell 49, 444-460.e9 (2019) doi:10.1016/j.devcel.2019.04.007 |
| Hu, K. H., Bruce, M. A., Liu, J. & Butte, M. J. Biochemical Stimulation of Immune Cells and Measurement of Mechanical Responses Using Atomic Force Microscopy. Current Protocols in Chemical Biology 11, e63 (2019) doi:10.1002/cpch.63 |
| Wall, A. A., Condon, N. D., Luo, L. & Stow, J. L. Rab8a localisation and activation by Toll-like receptors on macrophage macropinosomes. Philosophical Transactions of the Royal Society B: Biological Sciences 374, 20180151 (2018) doi:10.1098/rstb.2018.0151 |
| Condon, N. D. et al. Macropinosome formation by tent pole ruffling in macrophages. Journal of Cell Biology 217, 3873–3885 (2018) doi:10.1083/jcb.201804137 |
| Kashekodi, A. B., Meinert, T., Michiels, R. & Rohrbach, A. Miniature scanning light-sheet illumination implemented in a conventional microscope. Biomedical Optics Express 9, 4263 (2018) doi:10.1364/boe.9.004263 |
| Goltsev, Y. et al. Deep Profiling of Mouse Splenic Architecture with CODEX Multiplexed Imaging. Cell 174, 968-981.e15 (2018) doi:10.1016/j.cell.2018.07.010 |
| Kwak, B., Lee, Y., Lee, J., Lee, S. & Lim, J. Mass fabrication of uniform sized 3D tumor spheroid using high-throughput microfluidic system. Journal of Controlled Release 275, 201–207 (2018) doi:10.1016/j.jconrel.2018.02.029 |
| Jin, E. J. et al. Live Observation of Two Parallel Membrane Degradation Pathways at Axon Terminals. Current Biology 28, 1027-1038.e4 (2018) doi:10.1016/j.cub.2018.02.032 |
| Donnelly, S. K. et al. Rac3 regulates breast cancer invasion and metastasis by controlling adhesion and matrix degradation. Journal of Cell Biology 216, 4331–4349 (2017) doi:10.1083/jcb.201704048 |
| Ryan, D. P. et al. Automatic and adaptive heterogeneous refractive index compensation for light-sheet microscopy. Nature Communications 8, (2017) doi:10.1038/s41467-017-00514-7 |
| Singh, J. N., Nowlin, T. M., Seedorf, G. J., Abman, S. H. & Shepherd, D. P. Quantifying three-dimensional rodent retina vascular development using optical tissue clearing and light-sheet microscopy. Journal of Biomedical Optics 22, 1 (2017) doi:10.1117/1.jbo.22.7.076011 |
| McNamara, G., Difilippantonio, M., Ried, T. & Bieber, F. R. Microscopy and Image Analysis. Current Protocols in Human Genetics 94, (2017) doi:10.1002/cphg.42 |
| Thauland, T. J., Hu, K. H., Bruce, M. A. & Butte, M. J. Cytoskeletal adaptivity regulates T cell receptor signaling. Science Signaling 10, eaah3737 (2017) doi:10.1126/scisignal.aah3737 |
| A. Ahern, M., P. Black, C., J. Seedorf, G., D. Baker, C. & P. Shepherd, D. Hyperoxia impairs pro-angiogenic RNA production in preterm endothelial colony-forming cells. AIMS Biophysics 4, 284–297 (2017) doi:10.3934/biophy.2017.2.284 |
| Kotera, I. et al. Pan-neuronal screening in Caenorhabditis elegans reveals asymmetric dynamics of AWC neurons is critical for thermal avoidance behavior. eLife 5, (2016) doi:10.7554/elife.19021 |
| Huethorst, E. et al. Enhanced Human-Induced Pluripotent Stem Cell Derived Cardiomyocyte Maturation Using a Dual Microgradient Substrate. ACS Biomaterials Science & Engineering 2, 2231–2239 (2016) doi:10.1021/acsbiomaterials.6b00426 |
| Siegel, N., Lupashin, V., Storrie, B. & Brooker, G. High-magnification super-resolution FINCH microscopy using birefringent crystal lens interferometers. Nature Photonics 10, 802–808 (2016) doi:10.1038/nphoton.2016.207 |
| Miskolci, V., Hodgson, L. & Cox, D. Using Fluorescence Resonance Energy Transfer-Based Biosensors to Probe Rho GTPase Activation During Phagocytosis. in Methods in Molecular Biology 125–143 (Springer New York, 2016). doi:10.1007/978-1-4939-6581-6_9 |
| Siegel, N., Storrie, B., Bruce, M. & Brooker, G. CINCH (confocal incoherent correlation holography) super resolution fluorescence microscopy based upon FINCH (Fresnel incoherent correlation holography). in Quantitative Phase Imaging (eds. Popescu, G. & Park, Y.) (SPIE, 2015). doi:10.1117/12.2081319 |
| Lai, C. K. et al. Cell Fate Decisions in Malignant Hematopoiesis: Leukemia Phenotype Is Determined by Distinct Functional Domains of the MN1 Oncogene. PLoS ONE 9, e112671 (2014) doi:10.1371/journal.pone.0112671 |
| Perillo, E. P. et al. Enhanced 3D localization of individual RNA transcripts via astigmatic imaging. in Single Molecule Spectroscopy and Superresolution Imaging VII (eds. Enderlein, J., Gregor, I., Gryczynski, Z. K., Erdmann, R. & Koberling, F.) (SPIE, 2014). doi:10.1117/12.2038197 |
| Oreopoulos, J., Berman, R. & Browne, M. Spinning-disk confocal microscopy. in Methods in Cell Biology 153–175 (Elsevier, 2014). doi:10.1016/b978-0-12-420138-5.00009-4 |
| Eklund, A., Dufort, P., Forsberg, D. & LaConte, S. M. Medical image processing on the GPU – Past, present and future. Medical Image Analysis 17, 1073–1094 (2013) doi:10.1016/j.media.2013.05.008 |
| Zanella, R. et al. Towards real-time image deconvolution: application to confocal and STED microscopy. Scientific Reports 3, (2013) doi:10.1038/srep02523 |
| Bruce, M. A. & Butte, M. J. Real-time GPU-based 3D Deconvolution. Optics Express 21, 4766 (2013) doi:10.1364/oe.21.004766 |