Microvolution
Accelerate Your Discovery with Accuracy
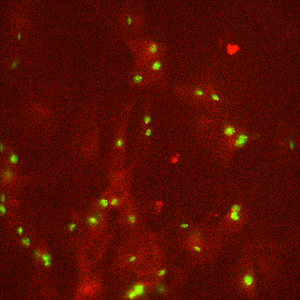
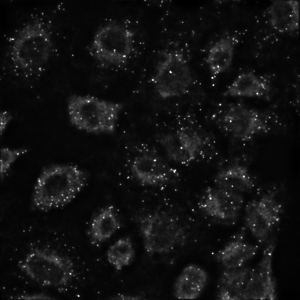
Widefield image of mitochondria. Example of excellent quality image restored from image captured dim light.
Single slice of 2048x2048x51 image, deconvolved in 8 seconds.
Image courtesy of: Dane Maxfield, Technical Instruments.
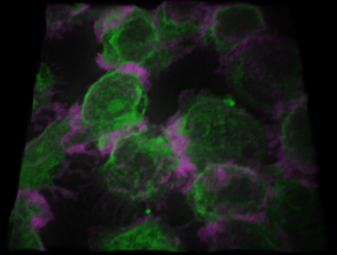
Instant SIM (iSIM) image, showing super resolution of raw image (208 nm) and further increase in resolution after deconvolution (117 nm).
Single slice of 1446x1131x25 image, deconvolved in 4.9 seconds.
Image courtesy of: Jim Paladino, BioVision.
Light sheet microscopy of Drosophila morphogenesis.
Single slice of 946x768x321 image, deconvolved in 22.5 seconds.
Image courtesy of: Dong-Yuan Chen in the Bilder lab, UC Berkeley.
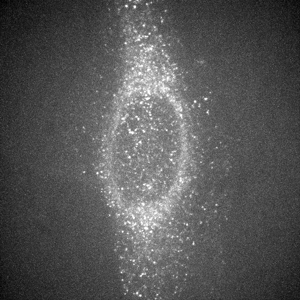
Spinning disk confocal image of endogenous ER marker.
Maximum intensity projection of 512x512x41 image, deconvolved in 0.7 seconds.
Image courtesy of: Cairn Research.
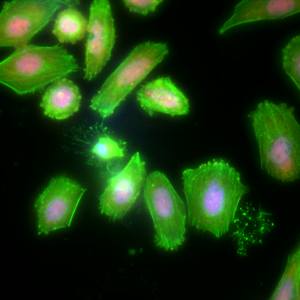
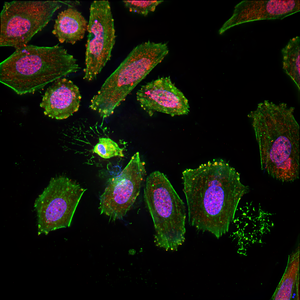
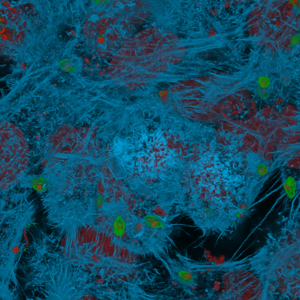
The human osteosarcoma cell line SaOS2 was subjected to differentiation conditions and stained to observe the regulation of an osteoblast transcription factor, RUNX2 (red), and a secreted protein, osteocalcin (green). Nuclei were counterstained with Hoechst 33342 (blue).A single osteocyte-like cell can be observed, characterized by a small nucleus and numerous, far-reaching cellular processes.
Maximum intensity projection of 2048x2048x32 image, 3 channels, deconvolved in 17 seconds total.
Image courtesy of: Dennis Hughes, MD, PhD, Assoc. Prof. of Pediatrics; Jared Mortus, Lead Researcher; Laurence Cooper, MD, PhD, PI; George McNamara, PhD, Sr Research Scientist; The Children’s Cancer Hospital, MD Anderson Cancer Center, Houston, TX.
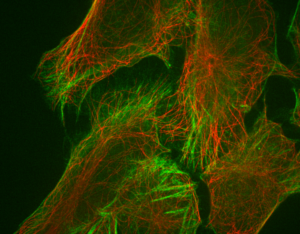
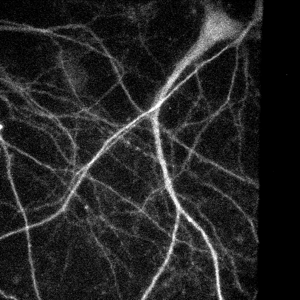
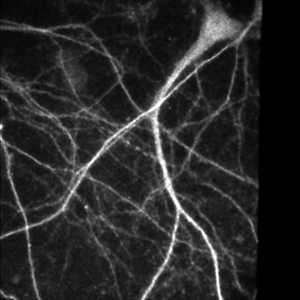
Spinning disk confocal image of neurites.
Single slice of 512x512x14 image, deconvolved in 0.7 seconds.
Image courtesy of: Huguenard lab, Stanford University.
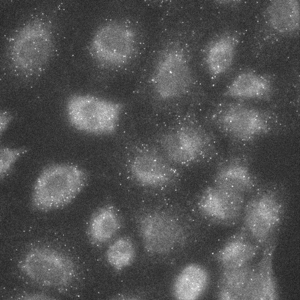
Widefield image of RNA FISH (fluorescence in-situ hybridization), labeling TOP1 (DNA topoisomerase 1).
Single slice of 1024x1024x39 image, deconvolved in 6 seconds.
Image courtesy of: Dane Maxfield, Technical Instruments.
HeLa cells and invading T. cruzi parasites, showing parasites releasing vesicles inside actin-rich cups formed at the surface of the HeLa cells. Top-down view of a 3D rendering in FluoRender.
1024x1024x34 3-channel image, deconvolved in 1.6 seconds.
Image courtesy of: Alexis Bonfim-Melo in the lab of Prof. Renato A. Mortara, Div. of Parasitology, Dept. of Microbiology, Immunology and Parasitology, Medical School of the Federal University of São Paulo.
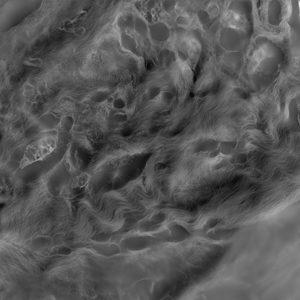
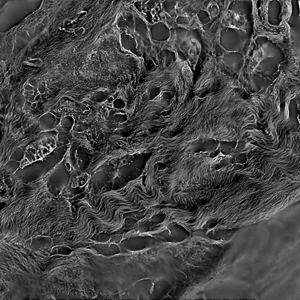
Collagen fibers in breast cancer tissue. The raw images were computed with using orientation-independent differential interference contrast (OI-DIC) microscopy (M. Shribak, “Quantitative orientation-independent DIC microscope with fast switching shear direction and bias modulation”, The Journal of the Optical Society of America A, vol.30, No.4, 769-782 (2013).)
Single slice of 1024x1024x51 image, blind deconvolved in 13.7 seconds.
Image courtesy of: Dr. Michael Shribak, Marine Biological Laboratory.